商品名称:QIAseq UPX 3' Targeted 96 index D (384)
QIAseq UPX 3' Targeted RNA Panels use unique molecular index (UMIs) and ultraplex (UPX) technology to enable accurate gene expression using 3' RNA-seq from single cells (1–1000), cell pellets and previously isolated total RNA (1 pg to 10 ng). Use our custom builder to design your custom panel for up to 1000 genes. UPX technology allows up to 384 plates of 384 samples (147,456 total samples) to be multiplexed together at one time. QIAseq UPX 3' Targeted Panels are available for 96 or 384 samples. For processing less than 96 samples at a time, use QIAseq UPX 3' Targeted RNA Panel (96) (cat. no. 333041), which is supplied with a breakable plate enabling processing of as few as 24 samples at one time for sequencing. The QIAseq UPX 3' Targeted RNA Panel (96-M) (cat. no. 333042) is for processing 96 samples in parallel 4 times (384 samples). The QIAseq UPX 3' Targeted RNA Panel (384) (cat. no. 333043) is for processing 384 samples simultaneously. QIAseq UPX 3' Targeted RNA Panels are only compatible with QIAseq UPX Targeted RNA Indices, which must be used during library construction for correct indexing.
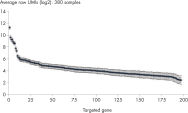 QIAseq UPX 3’ Targeted RNA Panel Workflow Exhibits Strong Linearity from 10 pg to 1 ng Total RNA.
QIAseq UPX 3’ Targeted RNA Panel Workflow Exhibits Strong Linearity from 10 pg to 1 ng Total RNA.


 ,
, 
 QIAseq UPX 3’ Targeted RNA Panel Workflow Exhibits Strong Linearity from 10 pg to 1 ng Total RNA.
QIAseq UPX 3’ Targeted RNA Panel Workflow Exhibits Strong Linearity from 10 pg to 1 ng Total RNA.